This is a guide to help you write correct PLN.
Change log:
| Date | Description |
|---|---|
2021-09-02 |
First version. |
2021-09-30 |
Finished sections on bridges to modified terminals and generic crosslinks. |
2022-02-16 |
Add self-contained versions of the PLN that has modifications. |
2023-03-09 |
Add section on residue extensions. |
1. PLN introduction
PLN stands for Protein Line Notation, the native format used by the Proteax software to define peptide and protein structures. It is a line entry format that covers plain linear and cyclic sequences, multiple chains, and bridged and crosslinked residues and terminals.
2. Basic format
In its simplest form a PLN entry is a string of uppercase single-letter residue codes with standard H- and -OH terminals added at the ends. An example of this would be the first chain of human insulin as shown below.
H-GIVEQCCTSICSLYQN-OH
Displaying this graphically as sequence and structure:
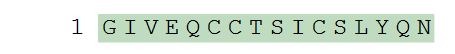
To put multiple chains into a PLN entry, the chains should be separated by periods. So to add the second chain of human insulin, add a period and then the second chain.
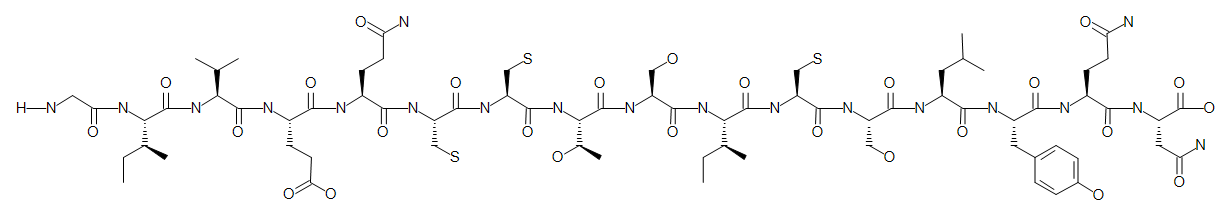
H-GIVEQCCTSICSLYQN-OH.H-FVNQHLCGSHLFYTPKT-OH
We now have the full sequence of human insulin. The sequence graphics uses alternating green and blue background colors to tell chains apart.

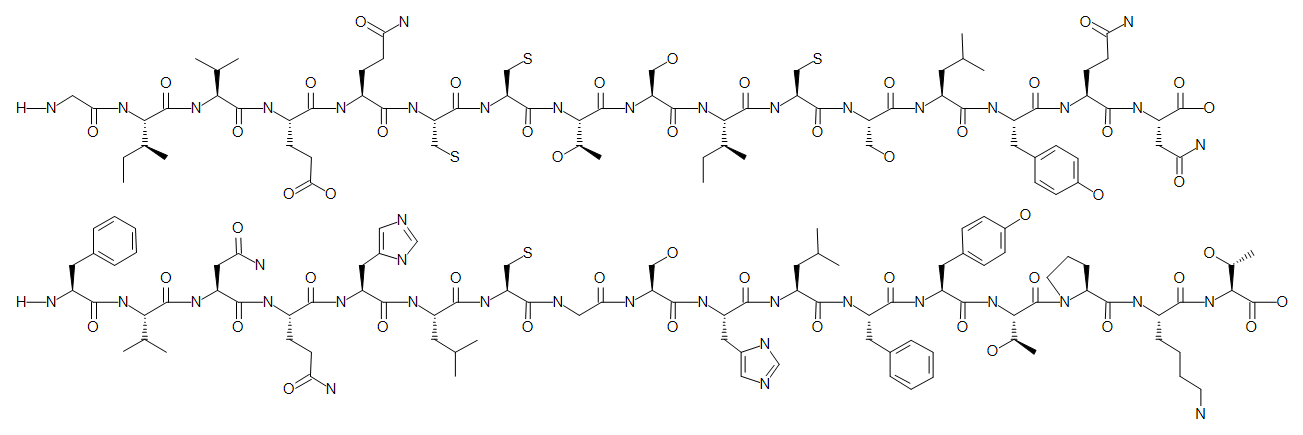
This is not quite the real structure of human insulin. We need to add disulfide bridges.
3. Disulfide bridges
A disulfide bridge is the simplest crosslink feature in PLN. It creates a disulfide bond linking two reactive sulfur atoms. As seleniumcysteine is a commonly occurring residue PLN accepts that disulfide bridges form between both sulfur and selenium atoms.
A disulfide bridge is defined in PLN by adding a unique number in parentheses after the residue or terminal that the disulfide bond should form to. Proteax will automatically find the reactive sulfur or selenium in the residue or terminal structure and create the disulfide bond.
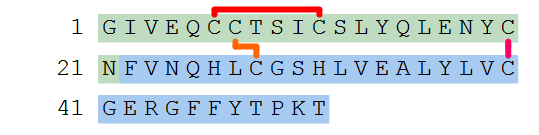
Human insulin has three disulfide bridges and here we number them 1 - 3. The numbers are used to pair up the residues/terminals that should be linked with disulfide bonds.
H-GIVEQC(1)C(2)TSIC(1)SLYQLENYC(3)N-OH.H-FVNQHLC(2)GSHLVEALYLVC(3)GERGFFYTPKT-OH
We now have an accurate structural definition of human insulin.
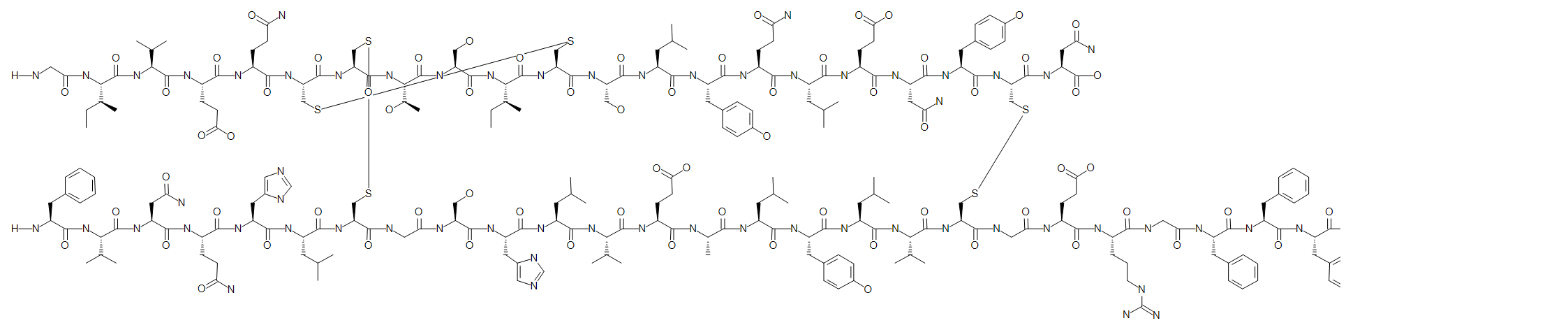
The numbers used do not carry any meaning but are solely used to uniquely link pairs of residues or terminals with disulfide bonds. We could use any pairs of numbers, so the following PLN will express the exact same sequence and structure as above.
H-GIVEQC(4)C(12)TSIC(4)SLYQLENYC(98)N-OH.H-FVNQHLC(12)GSHLVEALYLVC(98)GERGFFYTPKT-OH
3.1. Advanced disulfide bridges
Normal terminals cannot be linked via disulfide bridges as a bridge requires a pair of sulfur/selenium atoms. However, if you specify non-standard terminal structures that have a reactive sulfur or selenium, then you can create disulfide bridges from terminals. More about that in the section on modified terminals. *
4. Lactam cycles
A lactam cycle links a reactive nitrogen with an acid group, displacing a water molecule as shown below.
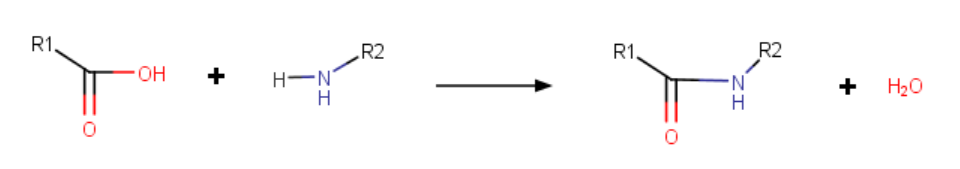
Lactam cycles are specified in PLN much like disulfide bridges. In the parentheses, add the keyword "lactam" or "cyclo" in front of the unique bridge number. An example is shown below.
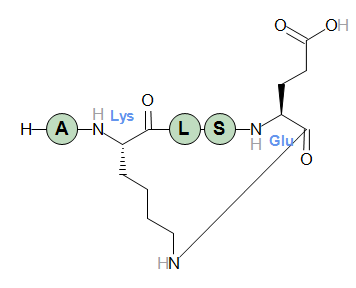
H-AE(cyclo1)DGIK(cyclo1)E-OH
The graphics of the structure has been switched to condensed mode so only structurally modified or crosslinked residues are shown as full structure. Standard residues are shown as "bubbles".

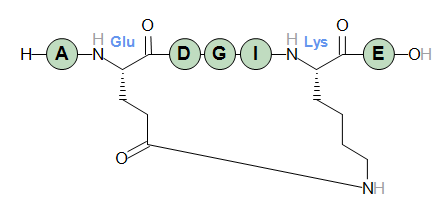
A terminal may also be the endpoint of a lactam cycle. Standard N-terminals provide a reactive amino group while C-terminals provide an acid group. The following two peptide examples demonstrate this. Note how the "(cyclo)" keyword replaces the "H-" or "-OH" standard terminal when the terminal is part of a bridge.
H-AK(cyclo1)LSE-(cyclo1)
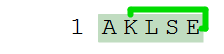
(cyclo)-AEDGIKE-(cyclo)

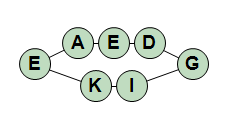
Note in the last example that when you cyclize a chain to be fully cyclic you do not need to add a numerical identifier after the "cyclo" keyword. You are still allowed to do it, but it is not necessary.
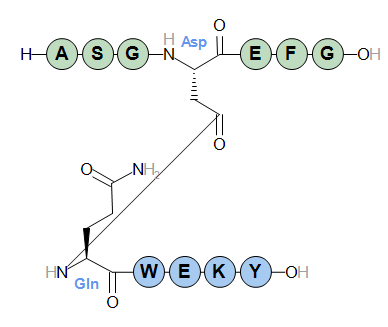
Finally, an example of linking two chains from a residue to a terminal, yielding a branched peptide.
H-ASGD(lactam1)EFG-OH.(lactam1)-QWEKY-OH

5. Thioether bridges
Another reactive crosslink formation is thioether bridges. This is performed via the "thio" keyword and builds an S-C link between a reactive sulfur site and a C-OH group (displacing the -OH).
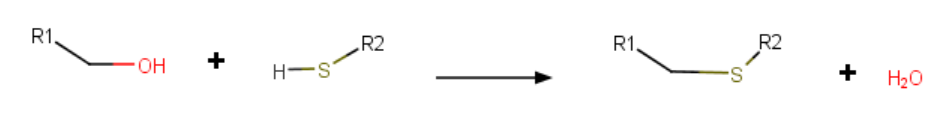
A simple example with a single thioether bridge.
H-ASC(thio1)ESFD(thio1)W-OH
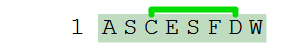
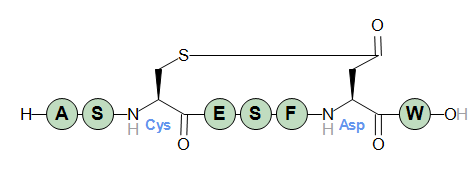
Thioether bridges can be used in PLN all the same places as lactam bridges, provided that the linked residues or terminals have reactive sites that match the thioether chemistry.
6. Modified residues and terminals
When a peptide is post-translationally or chemically modified, the standard one-letter residue codes and terminals do not faithfully represent the actual structure. In those cases, PLN annotations let you replace the standard residue codes and terminals with custom structures.
Custom structures that are used often will be stored in the Proteax modification database. Your modification database may however not have the structures used in the examples below, and that will make the PLN unreadable at your end. All PLN examples can therefore be found in self-contained format using inline modifications at the end of this document. This will let you copy/paste the PLN directly without having to register any additional custom structures in your modification database.
6.1. Modified structures
Non-standard residues or terminals are specified by name in square brackets.
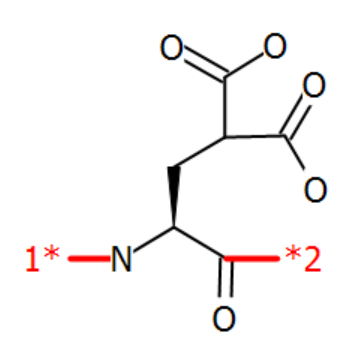
An example is the gamma-carboxy version of standard glutamate. An often-used abbreviation for gamma-carboxy-glutamate is "Gla" and in a default Proteax installation the "Gla" structure is already registered in its monomer database. The structure of "Gla" is shown below.
We can then write the PLN below to get a modified version of the first Glu/E residue in the sequence.
H-GIV[Gla]ASDE-OH
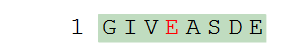
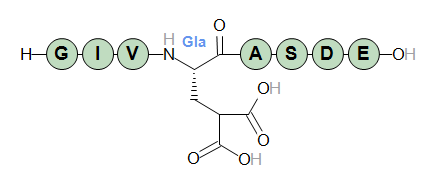
The modified "E" residue is highlighted in red in the sequence graphics, and expanded to full structure in the structure graphics.
In the same way, modified terminal structures can be specified by name. A default Proteax installation has the name "[NH2]" registered, representing an amidated terminal.
The sequence above with an amidated C-terminal will then be as follows
H-GIV[Gla]ASDE-[NH2]
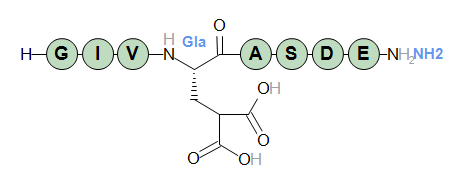
In the sequence graphics modified terminals are indicated by highlighted background stripes at the end of the chain.
6.2. D-forms
Structures can also be modified by applying a D-transform modifier. This takes the form of a "{d}" prefixed on any residue or terminal, standard or modified. The "{d}" transformation inverts all tetrahedral stereo centers in the affected structure.
The following example illustrates the usage. The
H-G{d}IV[Gla]ASD{d}[Gla]-OH
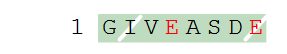
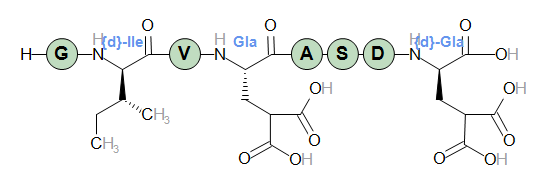
Residue 2 is marked as a D-form, residue 4 is modified, residue 8 is both modified and transformed to D-form. In the sequence graphics D-forms are marked with a white slash in the background color.
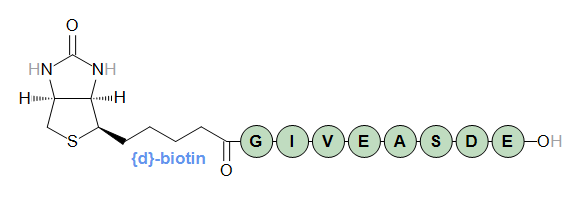
Modified terminals can also be D-transformed if they have active stereo centers. The "biotin" N-terminal structure is optically active.
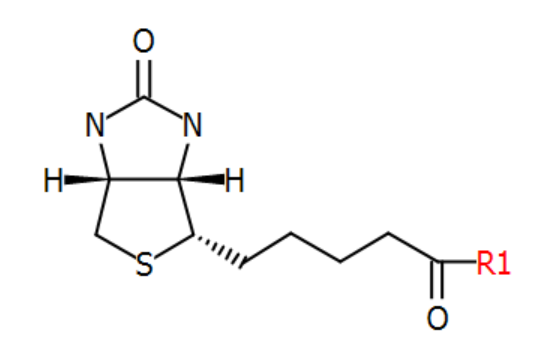
When D-transformed, its stereo configuration will be inverted. All stereo bonds are flipped from "up" to "down" and from "down" to "up".
{d}[biotin]-GIVEASDE-OH
7. Modifications and bridges
Bridges can be formed to modified residues and terminals, just like standard residues and terminals. Place the bridge parentheses after the modification name, like you would place it after a residue code or in place of a standard terminal.
In this section an "S-term" N-terminal structure containing a sulfur is used.
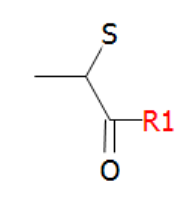
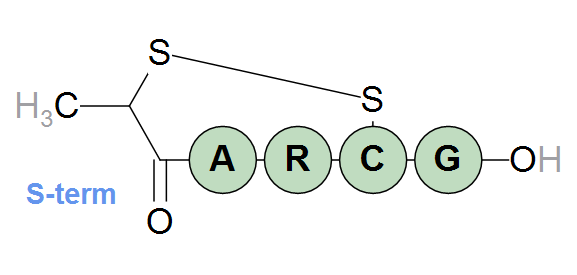
The sulfur of the terminal structure can be linked to a cysteine in the sequence through a normal disulfide bridge as in the example below.
[S-term](1)-ARC(1)G-OH
8. Generic crosslinks
Some complex structures cannot be expressed using ordinary sidechain modifications and standard reactive bridges. You may have sidechains with ambiguous reactive sites, multiple residues that are linked (three or more), or simply linkages that happen via other reactions than the three standard bridges that Proteax supports.
In that case you will want to use generic crosslinks.
8.1. Crosslinks linking two endpoints
An example is a stapled peptide from literature: Chimia 2013, 67, nr. 12, p. 902. The crosslink structure "staple" below is used to link two of the residues in the peptide. The two residue endpoints have their peptide bonds represented by R-group atom pairs R1-R2 and R3-R4. The purple-marked bond is the crossbond that connects the two endpoints. The crossbond will be stretched as-needed when the final structure is layed out.
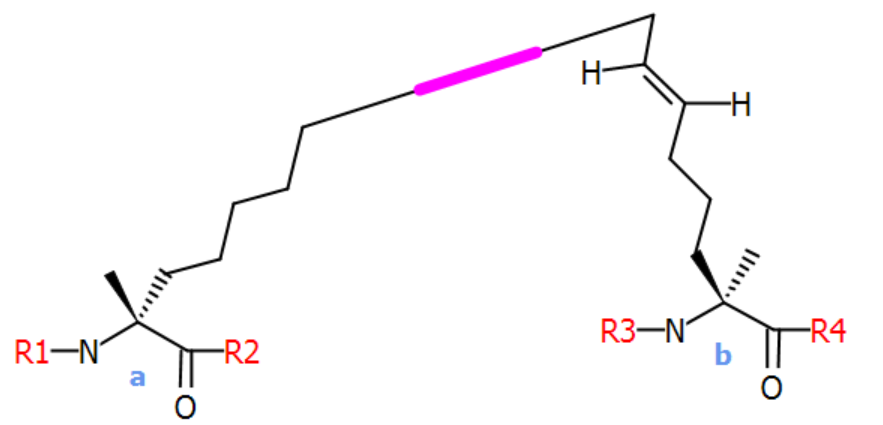
The full peptide sequence of the stapled peptide is shown below. The crosslink structure is referenced by its name "[staple]" at each residue endpoint. The crosslink is formed by adding a parenthesis with a crosslink number followed by an "@" and an endpoint specification - "a" or "b".
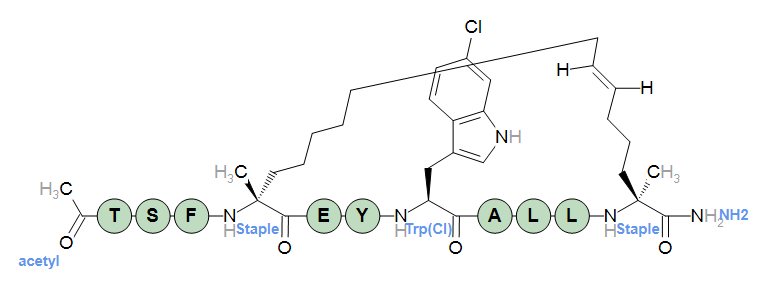
[acetyl]-TSF[staple](1@a)EY[Trp(Cl)]ALL[staple](1@b)-[NH2]
A crosslink endpoint can be a terminal as well. Using the "S-term" terminal structure from before, the following crosslink can be defined.
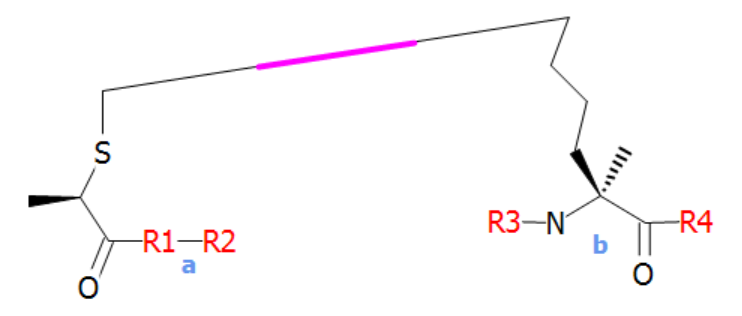
Example usage could be:
[N-term_res](1@a)-G[N-term_res](1@b)G-OH
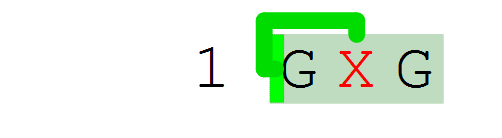

8.2. Crosslinks with cores
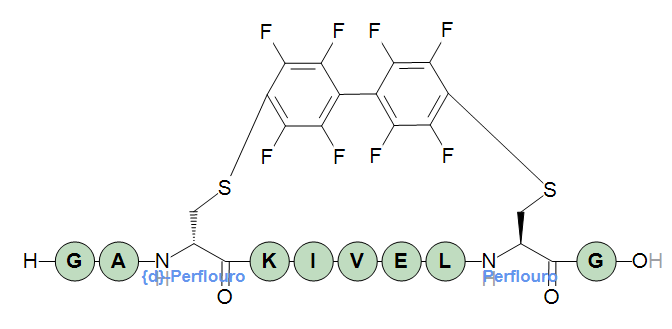
Crosslinks that only link two endpoints do not need to define a core structure. As soon as you link more than two endpoints a core structure is required, and in some cases it is practical to define a core structure anyway as in this crosslink that is a perflouroarene-based linker.
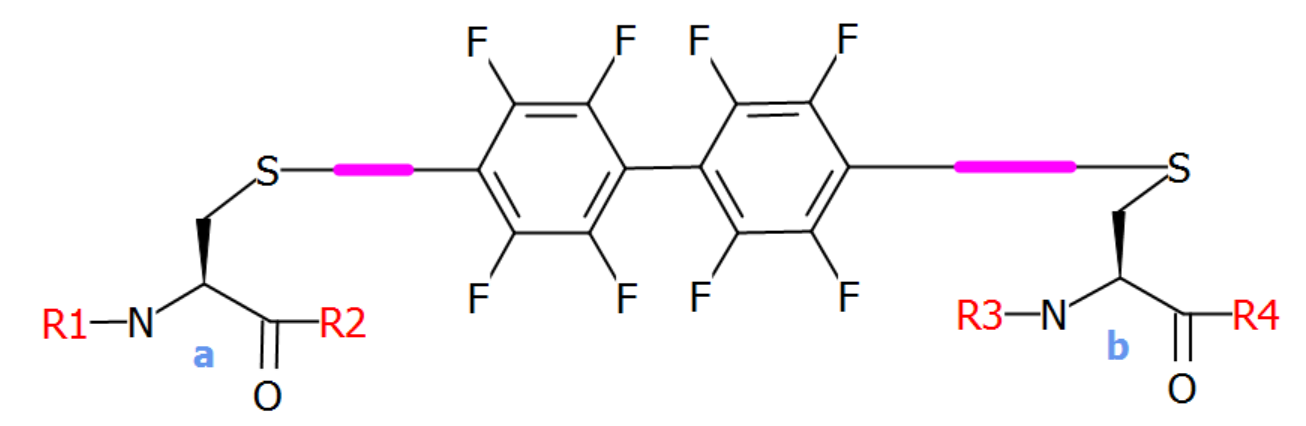
The crosslink can be used like this:
H-GA[perflouro](1@a)KIVEL[perflouro](1@b)G-OH
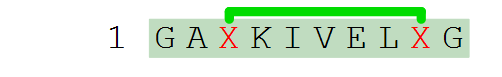
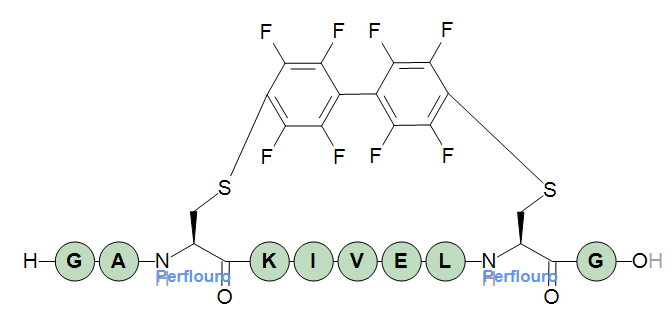
Endpoints can be stereo-inverted by applying a {d} transformation. The transformation will only applies to the endpoint structure until a crossbond is encountered. This means that endpoints do not affect each other, and a core structure will never be affected by any {d} transformations of endpoints. In the example below, the first cysteine endpoint has been {d} transformed which does not affect the second endpoint (nor the core, but that would have no effect anyway for this particular core).
H-GA{d}[perflouro](1@a)KIVEL[perflouro](1@b)G-OH
9. Residue extensions
Residue extensions let you attach terminal structures to residue sidechains. This way you can combine existing structures without having to register new residue structures.
The general format of a residue extension is
[RRR<A>{tx}X:TTT]
where
-
RRR is a residue name.
-
Either a standard one- or three-letter residue or the name of a modified residue.
-
-
A is an atom symbol defining the reactive site in the residue structure.
-
When multiple atoms match, the atom with the most free valences will be chosen as the reactive site.
-
-
TTT is a terminal name, with the prefixed X being "N" or "C" to fully qualify the name.
-
{tx} are optional commands that transform the terminal structure referenced by the TTT name.
-
Available commands are "d", "flipHorz", "flipVert", "rotate".
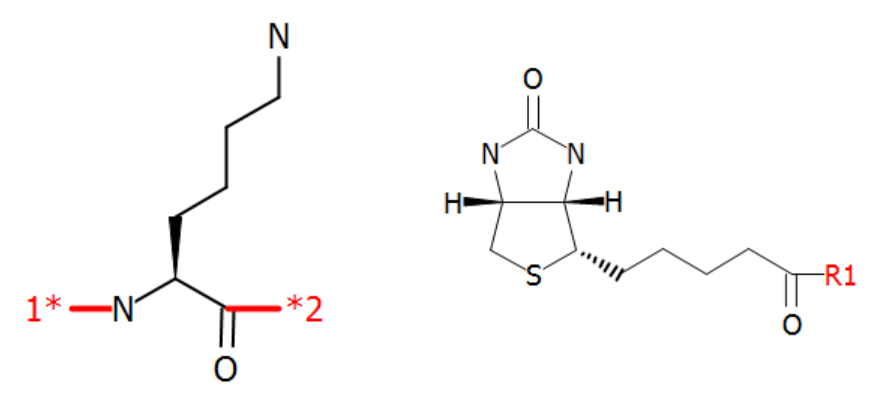
-
An example is biotinylation of lysine. This can be achieved by extending a standard lysine structure with an N-terminal biotin. These two structures are shown below.
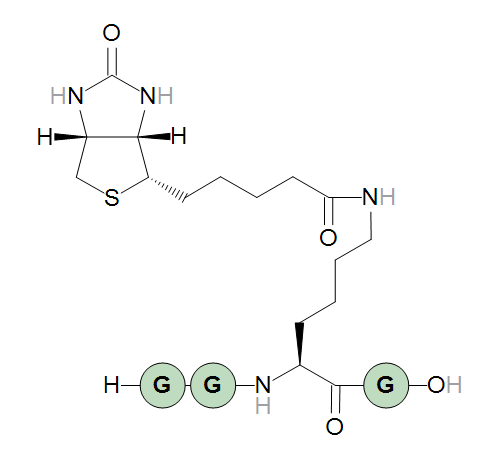
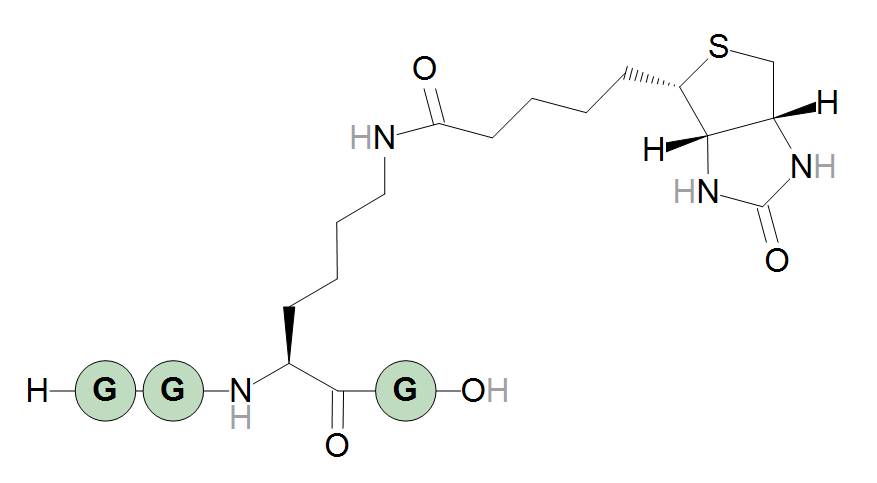
A sequence that generates a lysine extended with a biotinylation can then be:
H-GG[Lys<N>N:biotin]G-OH
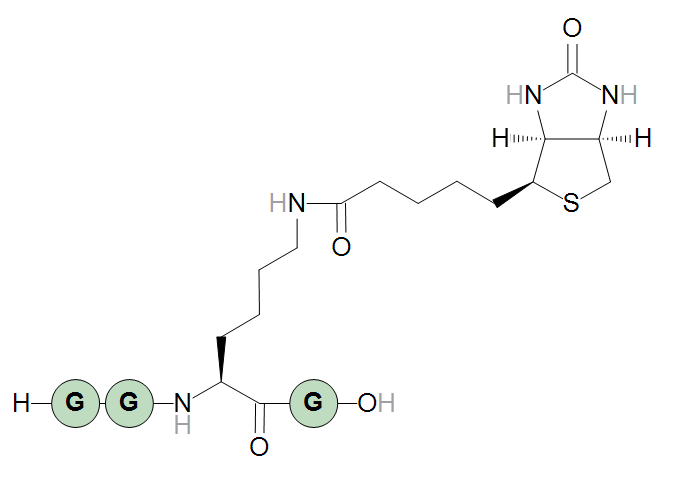
The optional terminal transforms can be used to flip the terminal structure’s stereochemistry (the "d" transform) or change the terminal structure’s layout (the remaining options). An example is flipping the horizontal layout of the biotin with the "flipHorz" transform.
H-GG[Lys<N>{flipHorz}N:biotin]G-OH
We can also flip the terminal vertically and rotate it 15 degrees.
H-GG[Lys<N>{flipHorz,flipVert,rotate15}N:biotin]G-OH
To rotate clockwise, use negative numbers e.g. "rotate-15".
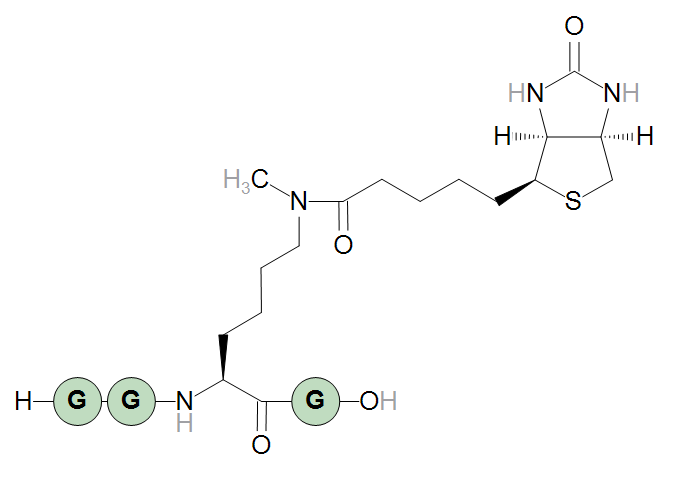
Finally, an example of biotinylation of a modified lysine, N6-methyllysine.
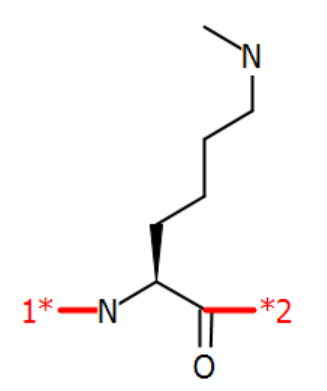
H-GG[N6-methyllysine<N>{flipHorz}N:biotin]G-OH
10. PLN examples in self-contained format
If you copy the below PLN examples from a PDF file you are unfortunately likely to see errors when pasting the text into the PLN editor. Some PDF readers convert regular spaces into special non-breaking-space characters and others remove hyphens from the end of a line. If you experience these issues, you can copy from the HTML version instead (use the second link to go straight to this section):
http://biochemfusion.com/doc/PLN_Guide/PLN_Guide.html
http://biochemfusion.com/doc/PLN_Guide/PLN_Guide.html#_pln_for_copying
[Gla] usage.
H-GIV[Gla]ASDE-OH inline-mod=E-residue,[Gla],C6H7N1O5,QkNGTREMC/8ODgBA/rwHj80PAFD+2QYfjBEAQP68Bg+HEQAg/qwIv8kPAHH+PQZfhhEAgf6OBs+BEQCh/ucGHz4TALL+QggPwQ8Asf7FCA+IEwCB/q0Gj4sUAGX+1AgviBQAnf6lCAQFGAABGAUGGAIDKAYHGAYIKAUJGAEEGQkKGAECGAkLKEEEAAECAho=
[Gla] usage with amidated C-terminal.
H-GIV[Gla]ASDE-[NH2] inline-mod=E-residue,[Gla],C6H7N1O5,QkNGTREMC/8ODgBA/rwHj80PAFD+2QYfjBEAQP68Bg+HEQAg/qwIv8kPAHH+PQZfhhEAgf6OBs+BEQCh/ucGHz4TALL+QggPwQ8Asf7FCA+IEwCB/q0Gj4sUAGX+1AgviBQAnf6lCAQFGAABGAUGGAIDKAYHGAYIKAUJGAEEGQkKGAECGAkLKEEEAAECAho= inline-mod=C-terminal,[NH2],H2N1,QkNGTRECAV8hBAB7/zAHvx0CAHv/MAABABhSAgEBGg==
[Gla] D-forms.
H-G{d}IV[Gla]ASD{d}[Gla]-OH
inline-mod=E-residue,[Gla],C6H7N1O5,QkNGTREMC/8ODgBA/rwHj80PAFD+2QYfjBEAQP68Bg+HEQAg/qwIv8kPAHH+PQZfhhEAgf6OBs+BEQCh/ucGHz4TALL+QggPwQ8Asf7FCA+IEwCB/q0Gj4sUAGX+1AgviBQAnf6lCAQFGAABGAUGGAIDKAYHGAYIKAUJGAEEGQkKGAECGAkLKEEEAAECAho=
N-terminal biotinylation.
{d}[biotin]-GIVEASDE-OH
inline-mod=N-terminal,[biotin],C10H15N2O2S1,QkNGTRESE4/c+/8e/xwQz5r9/y7/Qgb/WP//Hv9FBj8XAQAu/2sGf9UCAB7/cAa/kwQALv+VBv9RBgAe/5oGX08GAP7+Twi/G/r/Lv9CBh8T/f9N/5MGz6D6/03/kwYfGfr/bP/jBz+Y/f9s/+MHX9f7/33/Mga/1Pv/nf99CL8W//9N/6YBP534/03/fgGvVggAHv+aAAYHKAMEGAAIGAECFwEJGAQFGAgKGAABGAoLGAUGGAkMGAIDGAsNGAwNGAYRGA0OKAoJGAkPGQoQGVICEQEa
N-terminal with sulfur.
[S-term](1)-ARC(1)G-OH inline-mod=N-terminal,[S-term],C3H5O1S1,QkNGTREGBU8MAQCi/xYGrwkBAIH/3AifoP//uP/uBg+c/f+5/zMGj2gAANb/oxDvDwMAov8WAAAFGAACGAIDGAABKAIEGFICBQEa
Crosslinking in a stapled peptide.
[acetyl]-TSF[staple](1@a)EY[Trp(Cl)]ALL[staple](1@b)-[NH2] inline-mod=N-terminal,[acetyl],C2H3O1,QkNGTREEA+/nDQBs/q4Gj+sPAGz+rgAf5gwAiP6XBh/mDABQ/sYIAAIYAAEYAAMoUgIBARo= inline-mod=W-residue,[Trp(Cl)],C11H9Cl1N2O1,QkNGTREPEACjBQAuAD0HIIUCAB4AUQYwcQQAFAArBh86///d/9IHr/gAAO3/7wY/twIA3f/SBi+yAgC9/8II0PQAAA4AUwYghwIAPgC1BqB2BABIAJEG4OQEAGgAQAZAZAMAfgAcBqBzAQB0ABAGsAcBAFQAZgYA0QMAnQCdEQUGKAIAGAQHGQcBGAAJGAgJKAMEGAkKGAgBGAoLKAQFGAsMGAECKAwNKA0IGAsOGEEEAwEFAho= inline-mod=C-terminal,[NH2],H2N1,QkNGTRECAV8hBAB7/zAHvx0CAHv/MAABABhSAgEBGg== inline-mod=Crosslink,[Staple],C17H28N2O2,QkNGTTEbGn8F+f+9/hYHD8T6/83+Mwafgvz/vf4WBo99/P+d/gYIX//2/7z+7ADfg/7/vf4WAH+++f/p/lsGH8L7/+n+WwZ/s/3/8f7RBk84/v8R/xEGvykAABn/iAb/qwAAPf8DBj9BDQB9/6EGrwkOAMj+uAc/yA8A2P7VBs+GEQDI/rgGv4ERAKj+qAiPAwwAyP7MAA+IEwDI/swAT8YQAPT+6Qavwg4A9P7pBn9HDwAU/ykGT9gNACv/HAYfXQ4ASv9cBu/tDABh/08Gv2AQAEr/XAEvDAsAXP+XAQABGAEHFwIDKAcIGAAEGA0OGA8QKA4PGA0RGA8SGAgJGA4TFwIFGA4UGQkKGBQVGAECGBUWGAoLGBYXGBcYKAsMGBcZGAEGGRgaGAwYGFIIBAEFAhEDEgR4ARUa
N-terminally-linked crosslink.
[N-term_res](1@a)-G[N-term_res](1@b)G-OH inline-mod=Crosslink,[N-term_res],C11H18N1O2S1,QkNGTTETEj/c+/8S/wsGH10OAEr/XAafgvz/vf4WBo99/P+d/gYI34P+/73+FgB/hwAAvf4/AM8X+//T/vwGnxL5/9T+Mgbf3/v/8f6wEK8JDgDI/rgHP8gPANj+1QbPhhEAyP64Br+BEQCo/qgIjwMMAMj+zAAPiBMAyP7MAE/GEAD0/ukGr8IOAPT+6QZ/Rw8AFP8pBk/YDQAr/xwGAgQYBAUYAAEYAgYYBgcZAgMoBggYCAAYCQoYCwwoCgsYCQ0YCw4YCg8XChAZEBEYERIYEgEYUggEAQUCDQMOBHgBAho=
Crosslink with a core.
H-GA[perflouro](1@a)KIVEL[perflouro](1@b)G-OH inline-mod=Crosslink,[perflouro],C18H8F8N2O2S2,QkNGTTEkJY/UEQDA/j4HH5MTAND+Wwa/URUAwP4+Bp9MFQCg/i4Ib84PAMD+UgDvUhcAwP5SAF+PEwDs/pkGDz0VAAD/IRAPNAgA//7FBh83CQDj/v8GPzoLAOT+LgbPOQwAAP82Bm8yCwAc/x0GLzAJABv/yga/KgIA//4WBr8tAwDj/lEG3zAFAOP+fwZ/MAYA//6HBg8pBQAb/24G3yYDABv/Gwafffz//v7WEH8F+f+9/hYHD8T6/83+Mwafgvz/vf4WBo99/P+d/gYIX//2/7z+7ADfg/7/vf4WAB+7+v/p/tgGTyACADb/1wl/JgYAN/9/CR8uAgDH/lQJLzUGAMf+rQl/NwgAyP4CCZ8+DADI/l0JnykIADf/hgnfLwwAOP8uCRARKBESGBITKBMOGA4PKBEIGAAEGA4UGAIFGAkKGAoLKAsMGAwNKA0IGAgJKBUWGBYbGRcYKBsUGBUZGBcaGBYXGBMcGAsHGBIdGAEGGQ8eGAYHGBAfGAABGAkgGAIDKAohGAECGA0iGA8QGAwjGFIIBAMFBBkBGgJ4AhcHGg==
Crosslink with a core - endpoint (a) stereo-inverted.
H-GA{d}[perflouro](1@a)KIVEL[perflouro](1@b)G-OH
inline-mod=Crosslink,[perflouro],C18H8F8N2O2S2,QkNGTTEkJY/UEQDA/j4HH5MTAND+Wwa/URUAwP4+Bp9MFQCg/i4Ib84PAMD+UgDvUhcAwP5SAF+PEwDs/pkGDz0VAAD/IRAPNAgA//7FBh83CQDj/v8GPzoLAOT+LgbPOQwAAP82Bm8yCwAc/x0GLzAJABv/yga/KgIA//4WBr8tAwDj/lEG3zAFAOP+fwZ/MAYA//6HBg8pBQAb/24G3yYDABv/Gwafffz//v7WEH8F+f+9/hYHD8T6/83+Mwafgvz/vf4WBo99/P+d/gYIX//2/7z+7ADfg/7/vf4WAB+7+v/p/tgGTyACADb/1wl/JgYAN/9/CR8uAgDH/lQJLzUGAMf+rQl/NwgAyP4CCZ8+DADI/l0JnykIADf/hgnfLwwAOP8uCRARKBESGBITKBMOGA4PKBEIGAAEGA4UGAIFGAkKGAoLKAsMGAwNKA0IGAgJKBUWGBYbGRcYKBsUGBUZGBcaGBYXGBMcGAsHGBIdGAEGGQ8eGAYHGBAfGAABGAkgGAIDKAohGAECGA0iGA8QGAwjGFIIBAMFBBkBGgJ4AhcHGg==
Residue extension - Lys with N-terminal biotin.
H-GG[K<N>N:biotin]G-OH inline-mod=N-terminal,[biotin],C10H15N2O2S1,QkNGTRESE4/c+/8e/xwQz5r9/y7/Qgb/WP//Hv9FBj8XAQAu/2sGf9UCAB7/cAa/kwQALv+VBv9RBgAe/5oGX08GAP7+Twi/G/r/Lv9CBh8T/f9N/5MGz6D6/03/kwYfGfr/bP/jBz+Y/f9s/+MHX9f7/33/Mga/1Pv/nf99CL8W//9N/6YBP534/03/fgGvVggAHv+aAAYHKAMEGAAIGAECFwEJGAQFGAgKGAABGAoLGAUGGAkMGAIDGAsNGAwNGAYRGA0OKAoJGAkPGQoQGVICEQEa
Residue extension - N6-methyllysine with N-terminal biotin.
H-GG[N6-methyllysine<N>{flipHorz}N:biotin]G-OH
inline-mod=N-terminal,[biotin],C10H15N2O2S1,QkNGTRESE4/c+/8e/xwQz5r9/y7/Qgb/WP//Hv9FBj8XAQAu/2sGf9UCAB7/cAa/kwQALv+VBv9RBgAe/5oGX08GAP7+Twi/G/r/Lv9CBh8T/f9N/5MGz6D6/03/kwYfGfr/bP/jBz+Y/f9s/+MHX9f7/33/Mga/1Pv/nf99CL8W//9N/6YBP534/03/fgGvVggAHv+aAAYHKAMEGAAIGAECFwEJGAQFGAgKGAABGAoLGAUGGAkMGAIDGAsNGAwNGAYRGA0OKAoJGAkPGQoQGVICEQEa
inline-mod=K-residue,[N6-methyllysine],C7H14N2O1,QkNGTREKCQAAAAAAAAAHkL4BABAAHQYgfQMAAAAABh94AwDf//AIwLoBADAAgQZgdwMAQADSBtByAwBhACsGIC8FAHEAhgbwKgUAkQDVB2BsAwChAPIGAQQZAQIYBAUYAAEYBQYYAgMoBgcYBwgYCAkYQQQAAQICGg==